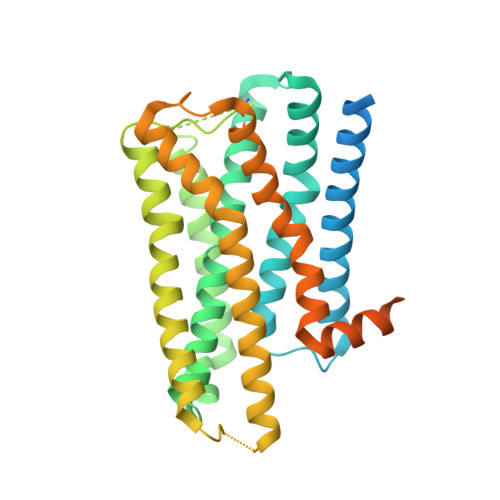
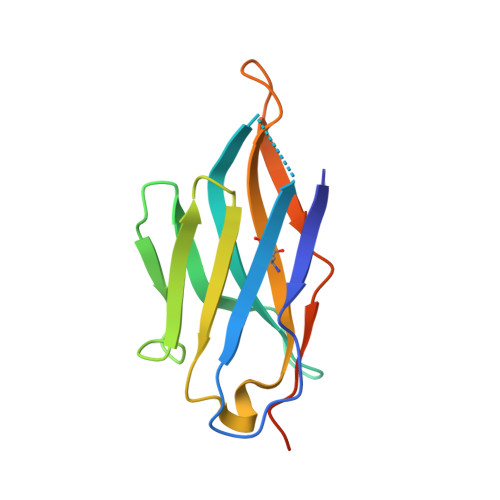
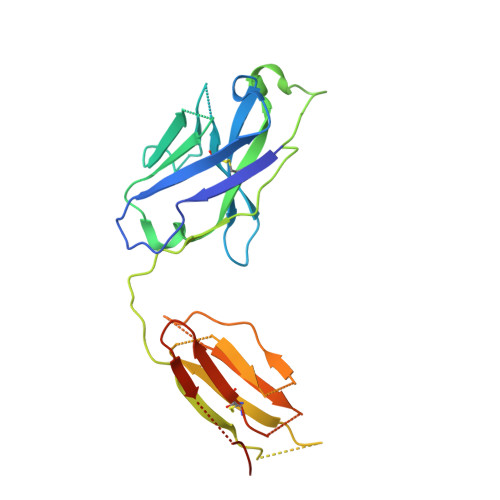
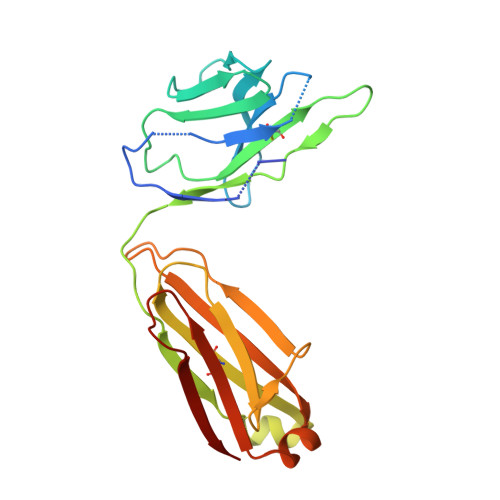
Structure determination of inactive-state GPCRs with a universal nanobody.
Robertson, M.J., Papasergi-Scott, M.M., He, F., Seven, A.B., Meyerowitz, J.G., Panova, O., Peroto, M.C., Che, T., Skiniotis, G.(2022) Nat Struct Mol Biol 29: 1188-1195
- PubMed: 36396979
- DOI: https://doi.org/10.1038/s41594-022-00859-8
- Primary Citation of Related Structures:
7UL2, 7UL3, 7UL4, 7UL5 - PubMed Abstract:
Cryogenic electron microscopy (cryo-EM) has widened the field of structure-based drug discovery by allowing for routine determination of membrane protein structures previously intractable. Despite representing one of the largest classes of therapeutic targets, most inactive-state G protein-coupled receptors (GPCRs) have remained inaccessible for cryo-EM because their small size and membrane-embedded nature impedes projection alignment for high-resolution map reconstructions. Here we demonstrate that the same single-chain camelid antibody (nanobody) recognizing a grafted intracellular loop can be used to obtain cryo-EM structures of inactive-state GPCRs at resolutions comparable or better than those obtained by X-ray crystallography. Using this approach, we obtained structures of neurotensin 1 receptor bound to antagonist SR48692, μ-opioid receptor bound to alvimopan, apo somatostatin receptor 2 and histamine receptor 2 bound to famotidine. We expect this rapid, straightforward approach to facilitate the broad exploration of GPCR inactive states without the need for extensive engineering and crystallization.
Organizational Affiliation:
Department of Molecular and Cellular Physiology, Stanford University School of Medicine, Stanford, CA, USA.